Human Genome Center, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan,
Kohji OKAMURA,![]() Yoichi YAMADA,
Yoichi YAMADA,![]() Yoshiyuki SAKAKI,
Yoshiyuki SAKAKI,![]() and Takashi ITO
and Takashi ITO![]()
Human Genome Center, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai,
Minato-ku, Tokyo 108-8639, Japan,![]() Division of Electrical and Computer Engineering, Graduate School of
Natural Science and Technology, Kanazawa University, 2-40-20 Kodatsuno,
Kanazawa 920-8667, Japan,
Division of Electrical and Computer Engineering, Graduate School of
Natural Science and Technology, Kanazawa University, 2-40-20 Kodatsuno,
Kanazawa 920-8667, Japan,![]() and Department of Computational Biology, Graduate School of Frontier
Sciences, University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa 277-8561,
Japan
and Department of Computational Biology, Graduate School of Frontier
Sciences, University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa 277-8561,
Japan![]()
Mouse Impact is the sole imprinted gene mapped to chromosome 18 to date. Despite its remarkable evolutionary conservation, human IMPACT was shown to escape genomic imprinting. Here we identified Hrh4 and Osbpl1 as the distal and proximal nearest neighbors of Impact, respectively, and found that both genes are expressed biallelically. Thus, in contrast with most imprinted genes, Impact fails to show apparent physical clustering with other imprinted genes. Since Impact not only lies in an intergenic region but also consists of 11 exons, it does not seem to be an imprinted gene generated by a retrotransposition. Hazardous effects of overexpressed Impact, a genomic segment containing paralogues of Hrh4 and Osbpl1 but not of Impact, and enhanced promoter activity in the mouse led us to propose an alternative model. This model assumes that segmental duplication followed by enhancement of the promoter activity in the lineage to mouse is responsible for the species-specific imprinting of Impact.
Impact; genomic imprinting; species-specific imprinting; segmental duplication; dosage compensation
Genomic imprinting is an epigenetic modification leading to exclusive
or highly skewed expression of a specific parental allele. The imprint
must be erased and established during gametogenesis, and the memory,
i.e. the parental origin, is maintained throughout the life of the
offspring. Although over 50 mammalian imprinted genes have been
discovered since 1991,![]() why and how they are imprinted remains largely unexplained.
why and how they are imprinted remains largely unexplained.
We previously developed a systematic screening for imprinted genes
using mRNA display PCR to identify a paternally expressed gene Impact,
which is preferentially expressed in brain to encode a protein of
unknown function but of remarkable evolutionary conservation.![]() It is the first and the sole imprinted gene mapped to mouse chromosome
18, which had been suggested to bear at least one imprinted gene by a
genetic study using mice with a Robertsonian translocation chromosome.
It is the first and the sole imprinted gene mapped to mouse chromosome
18, which had been suggested to bear at least one imprinted gene by a
genetic study using mice with a Robertsonian translocation chromosome.![]() Subsequently, we
Subsequently, we![]() and others
and others![]() isolated the cDNA for its human orthologue IMPACT and mapped it to human chromosome 18q11.2, a region syntenic to the mouse Impact locus.
isolated the cDNA for its human orthologue IMPACT and mapped it to human chromosome 18q11.2, a region syntenic to the mouse Impact locus.![]() Intriguingly, we revealed that human IMPACT is expressed from both alleles in all of the tissues examined so far.
Intriguingly, we revealed that human IMPACT is expressed from both alleles in all of the tissues examined so far.![]() It is thus suggested that the imprinting of Impact is species-specific.
It is thus suggested that the imprinting of Impact is species-specific.
Because most imprinted genes show close physical clustering, Impact
may provide a lead to the putative imprinted region on mouse chromosome
18. As for human, a parent-of-origin effect was observed in bipolar
affective disorder, and several groups provided evidence for a linkage
of the trait to the pericentric region of human chromosome 18.![]() Thus, although human IMPACT is expressed biallelically,
Thus, although human IMPACT is expressed biallelically,![]() it is of particular interest to identify its neighbors and examine their allelic expression status.
it is of particular interest to identify its neighbors and examine their allelic expression status.
In the present study, we analyzed allelic expression and methylation of the genes identified as the nearest neighbors of Impact as well as its human counterpart. Based on these observations, we propose a model for the evolution of imprinting of Impact.
BAC clones for mouse Impact (200P19 and 365M4) and human IMPACT (457A4 and 558E15) were identified by PCR screening as described previously.![]() The BAC clones 244P4 and 531H5 were obtained by additional rounds of
screening using sequence-tagged sites (STSs) developed from the
sequence of 365M4. The primers for a 230-bp STS, leading to the
identification of 244P4, were 5
The BAC clones 244P4 and 531H5 were obtained by additional rounds of
screening using sequence-tagged sites (STSs) developed from the
sequence of 365M4. The primers for a 230-bp STS, leading to the
identification of 244P4, were 5![]() -CAG ATG ACT AAC CCT GTT CA-3
-CAG ATG ACT AAC CCT GTT CA-3![]() and 5
and 5![]() -CCT AGG GTA TAA GCA ACT AC-3
-CCT AGG GTA TAA GCA ACT AC-3![]() .
The primers for a 319-bp STS, leading to the identification of 531H5 were 5
.
The primers for a 319-bp STS, leading to the identification of 531H5 were 5![]() -GCT TGT TAA CAT GTC AAC TTT C-3
-GCT TGT TAA CAT GTC AAC TTT C-3![]() and 5
and 5![]() -GCA GTT GAG ACA TTG CAT TAG T-3
-GCA GTT GAG ACA TTG CAT TAG T-3![]() .
.
Annotations for mouse, rat, and human genes were obtained from NCBI Map Viewer Web sites (http://www.ncbi.nlm.nih.gov/mapview/). Fugu Genome Browser on Ensembl Web sites (http://www.ensembl.org/Fugu_rubripes/) were used to identify pufferfish Impact and Hrh4 through tblastn search with the amino acid sequences of human IMPACT and HRH4 as queries.
For Northern hybridization, we used filters containing poly(A)![]() RNAs isolated from multiple tissues (Clontech).
Probes were purified by agarose gel electrophoresis and labeled with [
RNAs isolated from multiple tissues (Clontech).
Probes were purified by agarose gel electrophoresis and labeled with [![]() -
-![]() P]dCTP
(Amersham) using RadPrime DNA Labeling System (Invitrogen). The filters
hybridized with the probe were appropriately washed and exposed to
Imaging-Plates (Fuji Film) for analysis by Fuji BioImaging Analyzer
BAS2000 system. The 793-bp probe for mouse Osbpl1 was amplified from mouse brain cDNA with 5
P]dCTP
(Amersham) using RadPrime DNA Labeling System (Invitrogen). The filters
hybridized with the probe were appropriately washed and exposed to
Imaging-Plates (Fuji Film) for analysis by Fuji BioImaging Analyzer
BAS2000 system. The 793-bp probe for mouse Osbpl1 was amplified from mouse brain cDNA with 5![]() -TGC AGA AGG GCT CAA CAA TG-3
-TGC AGA AGG GCT CAA CAA TG-3![]() (forward) and 5
(forward) and 5![]() -AGT CCT CTT CCG ACT TGG AC-3
-AGT CCT CTT CCG ACT TGG AC-3![]() (reverse).
The 762-bp Osbpl1b-specific probe was amplified from mouse brain cDNA with 5
(reverse).
The 762-bp Osbpl1b-specific probe was amplified from mouse brain cDNA with 5![]() -GGT CGC TGA CAT CGA CTG TA-3
-GGT CGC TGA CAT CGA CTG TA-3![]() (forward) and 5
(forward) and 5![]() -GCT TGA GTC AGG TGT TTG CAT-3
-GCT TGA GTC AGG TGT TTG CAT-3![]() (reverse).
(reverse).
The Hpa II-McrBC PCR assay was performed as described.![]() Human peripheral blood samples used in this study were collected after
informed consent of 14 Japanese individuals in accordance with
ethical guidelines. Native genomic DNAs were digested with Hpa II or McrBC (New England Biolabs) for overnight, treated with phenol/chloroform, and precipitated by ethanol.
Subsequently, the digests were used as templates for PCR.
The PCR primers for the Osbpl1a CpG island are 5
Human peripheral blood samples used in this study were collected after
informed consent of 14 Japanese individuals in accordance with
ethical guidelines. Native genomic DNAs were digested with Hpa II or McrBC (New England Biolabs) for overnight, treated with phenol/chloroform, and precipitated by ethanol.
Subsequently, the digests were used as templates for PCR.
The PCR primers for the Osbpl1a CpG island are 5![]() -GCT ACA GCC AGG ATC CCT TA-3
-GCT ACA GCC AGG ATC CCT TA-3![]() (forward) and 5
(forward) and 5![]() -CCT GGG CTG GGT CTT GAA GA-3
-CCT GGG CTG GGT CTT GAA GA-3![]() (reverse).
Those for the Osbpl1b CpG island are 5
(reverse).
Those for the Osbpl1b CpG island are 5![]() -TGC TGC CGC CCC TCT TTC AC-3
-TGC TGC CGC CCC TCT TTC AC-3![]() (forward) and 5
(forward) and 5![]() -ACC CAC GCG GGC CGC TCT CA-3
-ACC CAC GCG GGC CGC TCT CA-3![]() (reverse), which were also used for direct sequencing.
Those for the HRH4 promoter were 5
(reverse), which were also used for direct sequencing.
Those for the HRH4 promoter were 5![]() -CCC TAG GAA TGT AAA GAC GAG-3
-CCC TAG GAA TGT AAA GAC GAG-3![]() (forward) and 5
(forward) and 5![]() -CCA GCC AGA CAA TTC TGA CA-3
-CCA GCC AGA CAA TTC TGA CA-3![]() (reverse).
Those for the OSBPL1A CpG island were 5
(reverse).
Those for the OSBPL1A CpG island were 5![]() -CAG GCT GCG CAA AGG TGA CT-3
-CAG GCT GCG CAA AGG TGA CT-3![]() (forward) and 5
(forward) and 5![]() -GCG GCC TCT GAA GAG CGG AT-3
-GCG GCC TCT GAA GAG CGG AT-3![]() (reverse).
Those for the OSBPL1B CpG island were 5
(reverse).
Those for the OSBPL1B CpG island were 5![]() -GGG AGT GCC AGC CAG AGT T-3
-GGG AGT GCC AGC CAG AGT T-3![]() (forward) and 5
(forward) and 5![]() -GGC ACG CAG CTG AAG ATC TG-3
-GGC ACG CAG CTG AAG ATC TG-3![]() (reverse).
(reverse).
Native genomic DNAs were denatured with 0.3 M sodium hydroxide at 37![]() C
for 15 min. The bisulfite (Sigma) solution, which was adjusted to
pH 5 with sodium hydroxide, and freshly prepared hydroquinone
(Sigma) were added to the denatured DNAs to the final concentrations of
3.2 M and 0.5 mM, respectively. The reaction mixes were
overlaid with 100
C
for 15 min. The bisulfite (Sigma) solution, which was adjusted to
pH 5 with sodium hydroxide, and freshly prepared hydroquinone
(Sigma) were added to the denatured DNAs to the final concentrations of
3.2 M and 0.5 mM, respectively. The reaction mixes were
overlaid with 100 ![]() l of mineral oil, and incubated at 55
l of mineral oil, and incubated at 55![]() C
overnight. The treated DNAs were purified by Wizard DNA Clean-Up System
(Promega) followed by desulfonation in 0.3 M sodium hydroxide for
15 min. Finally, the DNAs were precipitated by ethanol and used as
templates for PCR. Primers used for the upstream region of Hrh4 were 5
C
overnight. The treated DNAs were purified by Wizard DNA Clean-Up System
(Promega) followed by desulfonation in 0.3 M sodium hydroxide for
15 min. Finally, the DNAs were precipitated by ethanol and used as
templates for PCR. Primers used for the upstream region of Hrh4 were 5![]() -ATT TAG TGA TGG TTG GGG TTA GTT AAA T-3
-ATT TAG TGA TGG TTG GGG TTA GTT AAA T-3![]() (forward) and 5
(forward) and 5![]() -AAA TTC TAC CAA CCA CAT ACT AAA CCT A-3
-AAA TTC TAC CAA CCA CAT ACT AAA CCT A-3![]() (reverse).
Those for the promoter region of Hrh4 were 5
(reverse).
Those for the promoter region of Hrh4 were 5![]() -CCT ACA TCA CCT CTA AAC TTC TCT AAA A-3
-CCT ACA TCA CCT CTA AAC TTC TCT AAA A-3![]() (forward) and 5
(forward) and 5![]() -TTA AAA ATT TTT TTT TGA TTA TAA GAA GGA ATT-3
-TTA AAA ATT TTT TTT TGA TTA TAA GAA GGA ATT-3![]() (reverse).
Those for the Osbpl1b CpG island were 5
(reverse).
Those for the Osbpl1b CpG island were 5![]() -TCA CCC CCA CCT CCC CCA CCC AAA TCT-3
-TCA CCC CCA CCT CCC CCA CCC AAA TCT-3![]() (forward) and 5
(forward) and 5![]() -GGG TTA GGA GTA AGG AGG GTT TYG GGA A-3
-GGG TTA GGA GTA AGG AGG GTT TYG GGA A-3![]() (reverse).
The latter was also used for direct sequencing of the PCR products.
The PCR was performed with Platinum Taq DNA Polymerase (Invitrogen) which was complexed with antibody in order to inhibit polymerase activity until heat denaturation.
(reverse).
The latter was also used for direct sequencing of the PCR products.
The PCR was performed with Platinum Taq DNA Polymerase (Invitrogen) which was complexed with antibody in order to inhibit polymerase activity until heat denaturation.
Total RNAs were prepared by homogenizing frozen tissues in TRIZOL
Reagent (Invitrogen) followed by subsequent steps according to the
supplier's recommendations. We used QIAamp RNA Blood Mini Kits (Qiagen)
for the extraction from whole blood. Following DNaseI treatment,
reverse transcriptase reactions were performed using oligo-dT primer.
The PCR for Hrh4 was performed using primers 5![]() -AAT ATT GTC CTC ATT AGC TAC GAT CG-3
-AAT ATT GTC CTC ATT AGC TAC GAT CG-3![]() (forward) and 5
(forward) and 5![]() -TGG TTG CTT TGT CAC ACA AAG TAT CT-3
-TGG TTG CTT TGT CAC ACA AAG TAT CT-3![]() (reverse), and the products were directly sequenced with 5
(reverse), and the products were directly sequenced with 5![]() -TGT CTG TTC ACA ATT GTC CTT TCA AC-3
-TGT CTG TTC ACA ATT GTC CTT TCA AC-3![]() .
Those for Osbpl1a were the same as the one used for Northern blot hybridization.
The products were sequenced with the forward primer.
Those for Osbpl1b were 5
.
Those for Osbpl1a were the same as the one used for Northern blot hybridization.
The products were sequenced with the forward primer.
Those for Osbpl1b were 5![]() -GTG GTC GAG GAT CTG TTG AA-3
-GTG GTC GAG GAT CTG TTG AA-3![]() (forward) and 5
(forward) and 5![]() -CAG TTC ACA TCA GGA GGA TT-3
-CAG TTC ACA TCA GGA GGA TT-3![]() (reverse), and were sequenced with the forward primer.
The products were digested with Alu I overnight.
The corresponding genomic region was amplified with 5
(reverse), and were sequenced with the forward primer.
The products were digested with Alu I overnight.
The corresponding genomic region was amplified with 5![]() -CGT GGC AAT AAG CAG TGT TA-3
-CGT GGC AAT AAG CAG TGT TA-3![]() (forward) and 5
(forward) and 5![]() -AAG CTG AGC ATG TTC AAT CA-3
-AAG CTG AGC ATG TTC AAT CA-3![]() (reverse), and sequenced with 5
(reverse), and sequenced with 5![]() -GTG TGT GTA TTT ACT AAT G-3
-GTG TGT GTA TTT ACT AAT G-3![]() .
Those for HRH4 were 5
.
Those for HRH4 were 5![]() -TAC CTG TCA GTC TCA AAT GCT-3
-TAC CTG TCA GTC TCA AAT GCT-3![]() (forward) and 5
(forward) and 5![]() -GGG CAG ACC TGA TTC ATT TAG-3
-GGG CAG ACC TGA TTC ATT TAG-3![]() (reverse), and sequenced with 5
(reverse), and sequenced with 5![]() -ACT CAA CAT ACT GGG GTC TT-3
-ACT CAA CAT ACT GGG GTC TT-3![]() and 5
and 5![]() -CTT GGT TCT TGA GGA AAA CA-3
-CTT GGT TCT TGA GGA AAA CA-3![]() .
Those for OSBPL1 were 5
.
Those for OSBPL1 were 5![]() -GAA GAG GAC TGG AAG ACG AG-3
-GAA GAG GAC TGG AAG ACG AG-3![]() (forward) and 5
(forward) and 5![]() -TGA TAC TTA CAT GAG TGC AAC-3
-TGA TAC TTA CAT GAG TGC AAC-3![]() (reverse), and sequenced with 5
(reverse), and sequenced with 5![]() -CAG TTT TCT GCA GTC AGT ATC-3
-CAG TTT TCT GCA GTC AGT ATC-3![]() .
.
The RT-PCR was performed using oligo-dT primer and IMPACT primers 5![]() -GAT GAC TGT GCC AAA ATA TTT TGT ATT AG-3
-GAT GAC TGT GCC AAA ATA TTT TGT ATT AG-3![]() (forward), 5
(forward), 5![]() -TCT CTT ATT TTC TCC ACC CAC-3
-TCT CTT ATT TTC TCC ACC CAC-3![]() (reverse), or commercially available Gapd primers (Perkin Elmer) with the following thermal cycling parameters: 94
(reverse), or commercially available Gapd primers (Perkin Elmer) with the following thermal cycling parameters: 94![]() C for 60 sec followed by 20/25/30/50 cycles at 94
C for 60 sec followed by 20/25/30/50 cycles at 94![]() C for 20 sec, 58
C for 20 sec, 58![]() C for 40 sec, 72
C for 40 sec, 72![]() C for 10 sec, and a final extension at 72
C for 10 sec, and a final extension at 72![]() C for 180 sec.
We used 50 ng of total RNA isolated from B6 brain or 0.5-ng of human brain poly(A)
C for 180 sec.
We used 50 ng of total RNA isolated from B6 brain or 0.5-ng of human brain poly(A)![]() RNA (Clontech) for the assay.
Control PCR was performed using a 1-pg plasmid that has a 366-bp (B6) or 381-bp (human) cDNA fragment as the template.
RNA (Clontech) for the assay.
Control PCR was performed using a 1-pg plasmid that has a 366-bp (B6) or 381-bp (human) cDNA fragment as the template.
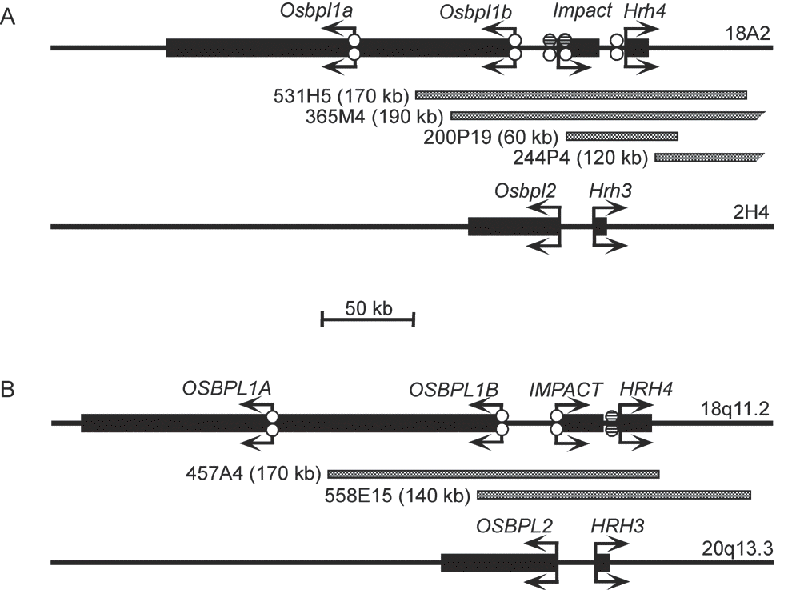
To obtain genomic clones containing the Impact/ IMPACT locus, we screened mouse and human bacterial artificial chromosome (BAC) libraries by PCR.![]() By subcloning and partial sequencing, we constructed BAC contigs covering the loci for mouse and human (Fig. 1).
Combining these and public data, we identified the histamine H
By subcloning and partial sequencing, we constructed BAC contigs covering the loci for mouse and human (Fig. 1).
Combining these and public data, we identified the histamine H![]() receptor gene (Hrh4/HRH4) and oxysterol binding protein-like 1 gene (Osbpl1/OSBPL1) as the distal and proximal nearest neighbors of Impact/IMPACT, respectively.
receptor gene (Hrh4/HRH4) and oxysterol binding protein-like 1 gene (Osbpl1/OSBPL1) as the distal and proximal nearest neighbors of Impact/IMPACT, respectively.
To assess whether mouse Hrh4 is imprinted, we performed an RT-PCR assay using F![]() hybrid mice generated by reciprocal crosses between Mus musculus molossinus JF1/Msf (JF) and M. musculus domesticus
C57BL/6J (B6). A G/A single nucleotide polymorphism (SNP) can be used
to distinguish the two parental alleles. Because this gene is
preferentially expressed in immune tissues, we extracted total RNAs
from the spleens of four different mice, JF, B6, and their reciprocal F
hybrid mice generated by reciprocal crosses between Mus musculus molossinus JF1/Msf (JF) and M. musculus domesticus
C57BL/6J (B6). A G/A single nucleotide polymorphism (SNP) can be used
to distinguish the two parental alleles. Because this gene is
preferentially expressed in immune tissues, we extracted total RNAs
from the spleens of four different mice, JF, B6, and their reciprocal F![]() hybrid mice, namely (JF
hybrid mice, namely (JF![]() B6)F
B6)F![]() and (B6
and (B6![]() JF)F
JF)F![]() .
All of the PCR primers for RNA analyses in this study were designed so
that each amplicon spans an exon-intron boundary for the prevention of
amplification from contaminated genomic DNAs. Direct sequencing of the
RT-PCR products showed that mouse Hrh4 is expressed biallelically (Fig. 2A).
We detected the expression of Hrh4 mRNA in the brain of some, but not all, of the hybrid mice.
Since Impact is highly expressed in brain, we used these mice to examine the allelic expression status of Hrh4 in the brain and found that it is expressed equally from both alleles (data not shown).
Thus, although Impact and Hrh4 are physically close (
.
All of the PCR primers for RNA analyses in this study were designed so
that each amplicon spans an exon-intron boundary for the prevention of
amplification from contaminated genomic DNAs. Direct sequencing of the
RT-PCR products showed that mouse Hrh4 is expressed biallelically (Fig. 2A).
We detected the expression of Hrh4 mRNA in the brain of some, but not all, of the hybrid mice.
Since Impact is highly expressed in brain, we used these mice to examine the allelic expression status of Hrh4 in the brain and found that it is expressed equally from both alleles (data not shown).
Thus, although Impact and Hrh4 are physically close (![]() 5 kb apart), their allelic expression status is distinct.
5 kb apart), their allelic expression status is distinct.
Next we examined the allelic methylation status of Hrh4.
In contrast to Impact, the Hrh4 gene lacks CpG islands.
We cloned and sequenced PCR products from bisulfite-treated DNA (Fig. 2B).
While we found that ![]() 1.4-kb
upstream region is hypermethylated in both spleen and brain, the
proximal promoter region was shown to escape methylation in spleen but
not in brain. We failed to find any clone of methylated allele from
spleen, and found no clone of unmethylated allele from brain. We thus
concluded that both alleles of the promoter region of Hrh4 escape methylation in spleen but are methylated in brain.
1.4-kb
upstream region is hypermethylated in both spleen and brain, the
proximal promoter region was shown to escape methylation in spleen but
not in brain. We failed to find any clone of methylated allele from
spleen, and found no clone of unmethylated allele from brain. We thus
concluded that both alleles of the promoter region of Hrh4 escape methylation in spleen but are methylated in brain.
The human OSBP genes were surveyed extensively,![]() and OSBPL1 was shown to produce two very different transcripts, the shorter one named OSBPL1A and the longer one called OSBPL1B.
Although mouse Osbp genes were recently identified,
and OSBPL1 was shown to produce two very different transcripts, the shorter one named OSBPL1A and the longer one called OSBPL1B.
Although mouse Osbp genes were recently identified,![]() Osbpl1b, whose promoter should be close to Impact, has not been reported.
To know whether Osbpl1b identified proximal to Impact
is indeed transcribed, we performed Northern blot hybridization using a
probe derived from the open reading frame (ORF) of mouse Osbpl1a.
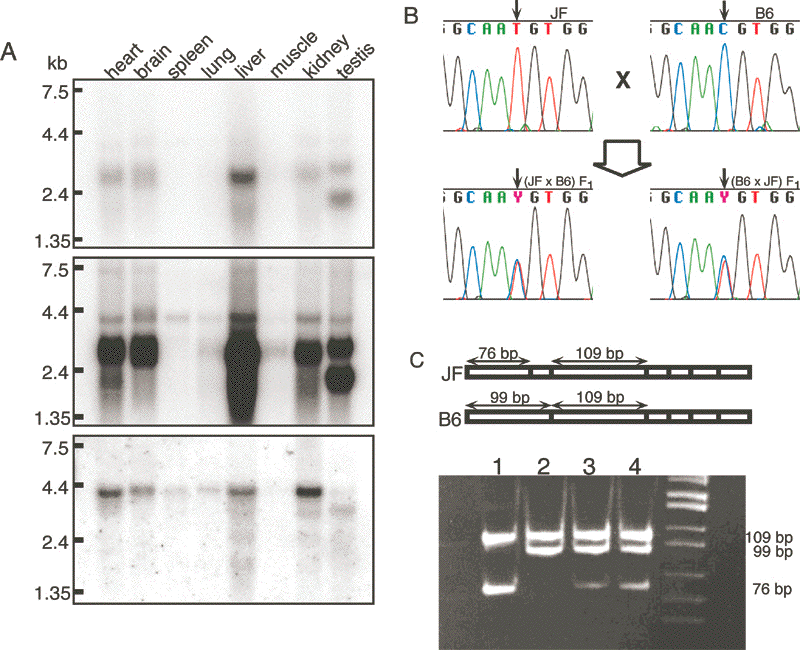
It detected a 3-kb mRNA in liver, heart, brain and kidney, whereas
testis contains an additional shorter transcript (Fig. 3A). Longer
exposure of the same blot revealed another band of 4-kb long, which may
be the transcript of Osbpl1b (Fig. 3A).
Osbpl1b, whose promoter should be close to Impact, has not been reported.
To know whether Osbpl1b identified proximal to Impact
is indeed transcribed, we performed Northern blot hybridization using a
probe derived from the open reading frame (ORF) of mouse Osbpl1a.
It detected a 3-kb mRNA in liver, heart, brain and kidney, whereas
testis contains an additional shorter transcript (Fig. 3A). Longer
exposure of the same blot revealed another band of 4-kb long, which may
be the transcript of Osbpl1b (Fig. 3A).
On the other hand, we noticed that a model reference sequence (GenBank accession no. XM_140455) was mapped to the promoter region of mouse Impact by NCBI RefSeq Project. We designed a forward primer in the model sequence and a reverse primer in Osbpl1a, and performed RT-PCR using mouse brain RNA as template. Consequently, we identified a long transcript that can encode a hypothetical protein consisting of 950 amino acids. The deduced primary sequence (GenBank accession no. AY536214) is 92% identical and 98% similar to human OSBPL1B. Northern blot hybridization using a probe derived from a region unique to this transcript did detect the 4-kb, but not the 3-kb, mRNA (Fig. 3A). These results indicate that the 4-kb band is indeed the mouse Osbpl1b mRNA, which is expressed in various tissues including spleen and lung where the shorter Osbpl1a mRNA is barely detected (Fig. 3A).
While mouse Osbpl1a and Osbpl1b transcripts are
expressed in various tissues (Fig. 3A), we used brain RNAs to
analyze allelic expression, because both forms of Osbpl1 as well as Impact are abundantly expressed in brain.![]() First, we designed primers around the 3
First, we designed primers around the 3![]() untranslated region (UTR) that is involved in both the Osbpl1a and Osbpl1b mRNAs.
However, since the quantity of the Osbpl1a transcript is
untranslated region (UTR) that is involved in both the Osbpl1a and Osbpl1b mRNAs.
However, since the quantity of the Osbpl1a transcript is ![]() 20 times larger than that of Osbpl1b
in brain (Fig. 3A), the latter is negligible in this assay. The
793-bp RT-PCR product obtained with these primers contains a T/C SNP
that allows us to discriminate the two parental alleles. Direct
sequencing of the RT-PCR products from the F
20 times larger than that of Osbpl1b
in brain (Fig. 3A), the latter is negligible in this assay. The
793-bp RT-PCR product obtained with these primers contains a T/C SNP
that allows us to discriminate the two parental alleles. Direct
sequencing of the RT-PCR products from the F![]() hybrid mice revealed the presence of both T and C alleles in the transcripts (Fig. 3B).
These results indicate that Osbpl1a is expressed biallelically (Fig. 3B).
hybrid mice revealed the presence of both T and C alleles in the transcripts (Fig. 3B).
These results indicate that Osbpl1a is expressed biallelically (Fig. 3B).
Next we designed Osbpl1b-specific primers, one in exon 3 and the other in exon 7, that give a 326-bp RT-PCR product.
This fragment has a T/C SNP that leads to a restriction fragment length polymorphisms (RFLP) when digested with Alu I (Fig. 3C).
The results of the RT-PCR RFLP assay showed that Osbpl1b was also expressed from both chromosomes.
Intriguingly, we noticed that the expression of the paternal allele was slightly stronger than that of the maternal one.
This inclination was more clearly shown by direct sequencing of the RT-PCR products of F![]() hybrid mice generated by reciprocal cross between B6 and JF (Fig. 3D).
Similar results were obtained with F
hybrid mice generated by reciprocal cross between B6 and JF (Fig. 3D).
Similar results were obtained with F![]() mice between JF and ICR (data not shown). To evaluate the skewed
expression quantitatively, we mixed the JF and B6 brain RNAs at various
ratios and subjected the mixed RNAs to RT-PCR followed by direct
sequencing. Comparing these results with those from reciprocal F
mice between JF and ICR (data not shown). To evaluate the skewed
expression quantitatively, we mixed the JF and B6 brain RNAs at various
ratios and subjected the mixed RNAs to RT-PCR followed by direct
sequencing. Comparing these results with those from reciprocal F![]() hybrid mice, we estimated that the amount of the expressed maternal
allele is approximately two thirds of the paternal one (Fig. 3E).
hybrid mice, we estimated that the amount of the expressed maternal
allele is approximately two thirds of the paternal one (Fig. 3E).
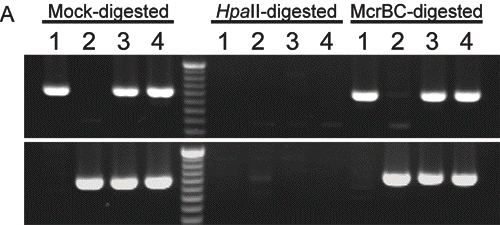
We also examined methylation status of CpG islands at the promoters of Osbpl1a and Osbpl1b by Hpa II-McrBC PCR assay.![]() In this assay, genomic DNA digested with Hpa II or McrBC was used as template for PCR. While the cleavage of CCGG site by Hpa II is blocked by methylation, McrBC selectively cuts methylated sequence (R
In this assay, genomic DNA digested with Hpa II or McrBC was used as template for PCR. While the cleavage of CCGG site by Hpa II is blocked by methylation, McrBC selectively cuts methylated sequence (R![]() CN
CN
![]() R
R![]() C).
C).![]() Hence, PCR products amplified from Hpa II- and McrBC-digested DNA are derived from hypermethylated and hypomethylated alleles, respectively.
As shown in Fig. 4A, both B6 and JF alleles of the CpG island at the Osbpl1b promoter were amplified from the McrBC-treated DNA but not from the Hpa
II-digested DNA, strongly suggesting that both alleles of this island
escape methylation. This notion was further reinforced by the results
of bisulfite sequencing, in which all of the CpGs were converted to
TpGs (Fig. 4B), indicating that the CpGs were modified to UpGs
upon bisulfite treatment as they had escaped methylation. Hence, we
cannot attribute the skewed expression of Osbpl1b to differential DNA methylation.
Since the promoter of Ospbl1b, which shows skewed expression, is much closer to Impact than that of Osbpl1a, which supports even biallelic expression, it is conceivable that an allele-specific difference in the chromatin structure of Impact somehow affects the function of the former.
We also obtained results indicating that neither allele of Osbpl1a promoter is methylated (data not shown).
We thus conclude that both alleles of the two CpG islands of Osbpl1 gene escape methylation.
Hence, PCR products amplified from Hpa II- and McrBC-digested DNA are derived from hypermethylated and hypomethylated alleles, respectively.
As shown in Fig. 4A, both B6 and JF alleles of the CpG island at the Osbpl1b promoter were amplified from the McrBC-treated DNA but not from the Hpa
II-digested DNA, strongly suggesting that both alleles of this island
escape methylation. This notion was further reinforced by the results
of bisulfite sequencing, in which all of the CpGs were converted to
TpGs (Fig. 4B), indicating that the CpGs were modified to UpGs
upon bisulfite treatment as they had escaped methylation. Hence, we
cannot attribute the skewed expression of Osbpl1b to differential DNA methylation.
Since the promoter of Ospbl1b, which shows skewed expression, is much closer to Impact than that of Osbpl1a, which supports even biallelic expression, it is conceivable that an allele-specific difference in the chromatin structure of Impact somehow affects the function of the former.
We also obtained results indicating that neither allele of Osbpl1a promoter is methylated (data not shown).
We thus conclude that both alleles of the two CpG islands of Osbpl1 gene escape methylation.
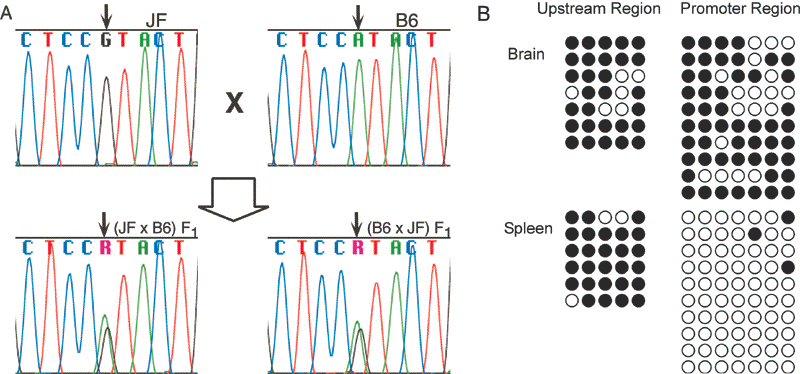
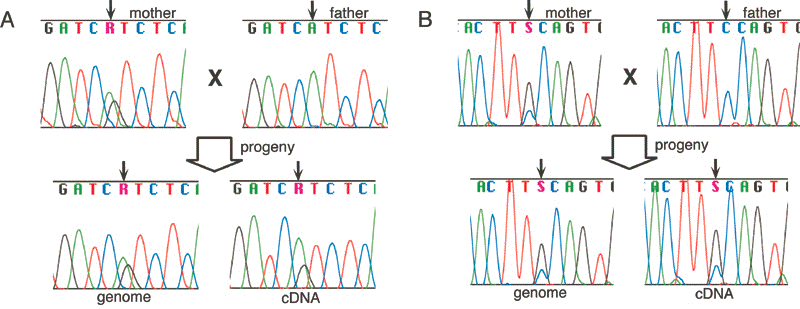
While we previously reported biallelic expression of human IMPACT,![]() it would be of importance to assess the imprinting status of adjacent genes.
To examine the imprinting status of human HRH4,
we searched polymorphisms in the transcribed region. Fortunately, we
found a G/A SNP and an informative Japanese family. This SNP alters an
amino acid residue from histidine to arginine. We performed RT-PCR
using RNA isolated from peripheral blood cells as the template. The
products were directly sequenced (Fig. 5A). The result shows that
the cDNA contained both maternal and paternal alleles, although
expression of the latter may be slightly higher than the former.
Because there is no CpG island in the HRH4 locus, we targeted the promoter region for DNA methylation analysis.
The result of Hpa
II-McrBC PCR assay suggests that both chromosomes are methylated (data
not shown). We thus assume it as a non-imprinted gene, although we
cannot completely exclude a possibility of loosely imprinted expression
of HRH4 in other tissues and other individuals.
it would be of importance to assess the imprinting status of adjacent genes.
To examine the imprinting status of human HRH4,
we searched polymorphisms in the transcribed region. Fortunately, we
found a G/A SNP and an informative Japanese family. This SNP alters an
amino acid residue from histidine to arginine. We performed RT-PCR
using RNA isolated from peripheral blood cells as the template. The
products were directly sequenced (Fig. 5A). The result shows that
the cDNA contained both maternal and paternal alleles, although
expression of the latter may be slightly higher than the former.
Because there is no CpG island in the HRH4 locus, we targeted the promoter region for DNA methylation analysis.
The result of Hpa
II-McrBC PCR assay suggests that both chromosomes are methylated (data
not shown). We thus assume it as a non-imprinted gene, although we
cannot completely exclude a possibility of loosely imprinted expression
of HRH4 in other tissues and other individuals.
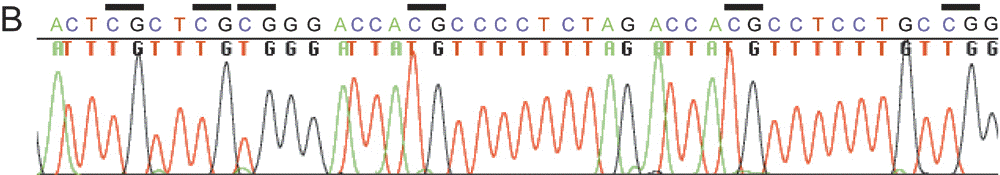
As for human OSBPL1, we found two Japanese families that are informative on a C/G SNP in the 3![]() UTR which is transcribed not only as OSBPL1A but also as OSBPL1B.
We extracted total RNAs from their blood cells and performed allelic
expression analysis. The results showed that they are expressed from
both alleles, although we could not distinguish the long and short
transcripts (Fig. 5B). The locations of the two CpG islands of OSBPL1 are conserved between mouse and human.
We performed DNA methylation analysis for the two islands by Hpa II-McrBC PCR assay.
It demonstrated that both islands escape methylation biallelically: they are usual unmethylated CpG islands (data not shown).
We thus concluded that both OSBPL1A and OSBPL1B are not imprinted.
UTR which is transcribed not only as OSBPL1A but also as OSBPL1B.
We extracted total RNAs from their blood cells and performed allelic
expression analysis. The results showed that they are expressed from
both alleles, although we could not distinguish the long and short
transcripts (Fig. 5B). The locations of the two CpG islands of OSBPL1 are conserved between mouse and human.
We performed DNA methylation analysis for the two islands by Hpa II-McrBC PCR assay.
It demonstrated that both islands escape methylation biallelically: they are usual unmethylated CpG islands (data not shown).
We thus concluded that both OSBPL1A and OSBPL1B are not imprinted.
As described above, it is unlikely that the locus around Impact/IMPACT forms a clear imprinted cluster.
Hence we may assume Impact as a putative solitary imprinted gene.
Some genes have been reported as solitary imprinted genes, e.g.
U2af1-rs1,![]() Nnat,
Nnat,![]() Nap1l5, and Peg13.
Nap1l5, and Peg13.![]() These are located within singular introns of other nonimprinted genes, Murr1, Blcap, Herc3, and KIAA1882,
respectively. Since these imprinted genes tend to have fewer and
smaller introns, retrotransposition has been considered a common
mechanism for the formation of such solitary ones. It is intriguing to
note that all of these genes presumably generated by retrotransposition
are paternally expressed like Impact.
However, these genes and Impact display remarkable differences in their genomic organization.
First, Impact has at least 10 introns and is much longer than other solitary imprinted genes.
These are located within singular introns of other nonimprinted genes, Murr1, Blcap, Herc3, and KIAA1882,
respectively. Since these imprinted genes tend to have fewer and
smaller introns, retrotransposition has been considered a common
mechanism for the formation of such solitary ones. It is intriguing to
note that all of these genes presumably generated by retrotransposition
are paternally expressed like Impact.
However, these genes and Impact display remarkable differences in their genomic organization.
First, Impact has at least 10 introns and is much longer than other solitary imprinted genes.![]() Second, Impact has no homologous genes in the genome except several processed pseudogenes in human.
Third, Impact does not reside within other genes but lies between two distinct genes, Hrh4 and Osbpl1.
These observations indicate that Impact is a unique solitary imprinted gene that was not generated by a retrotranspositional event.
Second, Impact has no homologous genes in the genome except several processed pseudogenes in human.
Third, Impact does not reside within other genes but lies between two distinct genes, Hrh4 and Osbpl1.
These observations indicate that Impact is a unique solitary imprinted gene that was not generated by a retrotranspositional event.
When considering non-retrotranspositional origin of Impact, a genomic segment attracted our attention. The segment resides on human chromosome 20q13.3 and contains both HRH3, displaying the highest homology with HRH4, and OSBPL2, the closest paralogue of OSBPL1, in a head-to-head orientation similar to the IMPACT locus on chromosome 18q11.2 (Fig. 1). Moreover, OSBPL1 and OSBPL2 are flanked by LAMA3 and its paralogue LAMA5, respectively, in a tail-to-tail manner. It thus seems that these were generated by duplication. However, the segment on chromosome 20 lacks any paralogue of IMPACT. Since retrotranspositional insertion of IMPACT into the segment on chromosome 18 is highly unlikely and a syntenic fragment containing Hrh4 and Impact can be found even in the puffer fish genome (data not shown), it seems that the duplicated copy of IMPACT is deleted from the segment on chromosome 20.
In this context, it is intriguing to note a recently proposed
hypothesis that underscores a role for duplications in the evolution of
imprinting.![]() According to this hypothesis, duplication leads to monoallelic
expression and subsequently imprinted expression of the resulting
paralogues for a simple basis of dosage compensation. This model is
based on their observation that the paralogues of imprinted genes are
often in close vicinity to other imprinted genes. This is true for the Osbpl1-Hrh4 locus, because Osbpl5,
According to this hypothesis, duplication leads to monoallelic
expression and subsequently imprinted expression of the resulting
paralogues for a simple basis of dosage compensation. This model is
based on their observation that the paralogues of imprinted genes are
often in close vicinity to other imprinted genes. This is true for the Osbpl1-Hrh4 locus, because Osbpl5,![]() a paralogue of Osbpl1, and Htr2a,
a paralogue of Osbpl1, and Htr2a,![]() another amine receptor similar to Hrh4, are maternally expressed genes on mouse chromosomes 7 and 14, respectively.
another amine receptor similar to Hrh4, are maternally expressed genes on mouse chromosomes 7 and 14, respectively.
We assume that an ancient segment containing Hrh-Impact-Osbpl-Lama was duplicated. However, since the increased dosage of Impact was hazardous, one of the copies was deleted from the genome, leaving a segment that eventually evolved to Hrh3-Osbpl2-Lama5.
Indeed, we previously demonstrated that overexpression of Xenopus homologue of Impact in oocyte inhibits normal embryonic development.![]() We also showed that the overexpression of YIH1, the yeast homologue of Impact, confers growth defect under amino acid starvation stress.
We also showed that the overexpression of YIH1, the yeast homologue of Impact, confers growth defect under amino acid starvation stress.![]() This may be the reason why only a single copy of Impact was retained in the vertebrate genomes, which, on the other hand, contain multiple paralogues of Hrh4 and Osbpl1.
This may be the reason why only a single copy of Impact was retained in the vertebrate genomes, which, on the other hand, contain multiple paralogues of Hrh4 and Osbpl1.
If the deletion of a duplicated copy of Impact restored the original level of gene dosage, it does not have to be imprinted. Indeed, we found that Impact is not imprinted in most vertebrates including frog, pig, monkey and human (in preparation). Then, why is Impact imprinted in mouse? One possibility is that the promoter activity was enhanced in the lineage leading to mouse. To control its dosage, mouse might exploit monoallelic expression, which eventually led to imprinted expression.
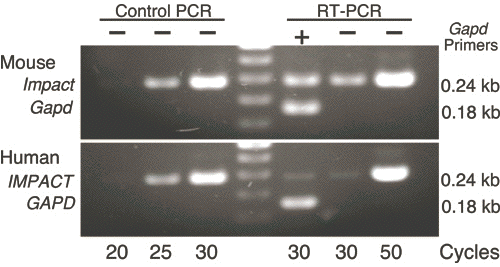
To examine whether the promoter activity is indeed enhanced in mouse, we compared the expression level of Impact and IMPACT by an RT-PCR assay using a single common primer pair for amplification of both genes. Although the primer sequences are derived from human IMPACT and not identical to the sequence of mouse Impact, the primer pair can amplify the latter as efficiently as the former (Fig. 6). The results of the assay clearly indicated that the expression of mouse Impact is much stronger than that of human IMPACT (Fig. 6). Similarly, our preliminary analysis indicated that the expression of Impact is significantly higher in rat and rabbit, both bearing imprinted Impact, than in monkey and pig, both bearing non-imprinted Impact (manuscript in preparation).
Based on these observations, we assume that the species-specific imprinting of Impact
evolved as a consequence of adaptations to increased gene dosage, which
was first induced by segmental duplication and next by enhancement of
the promoter activity in the lineage to rodents and rabbit. While the
mechanism leading to the enhanced promoter activity remains elusive, it
is intriguing to note the promoter sequence is considerably diverged
between mouse and human: the human promoter, but not the mouse one,
constitutes a conventional nonmethylated CpG island.![]() Furthermore, the first intron of the mouse gene, but not its human
counterpart, has a differentially methylated CpG island containing
characteristic tandem repeats, which are often found near imprinted
genes.
Furthermore, the first intron of the mouse gene, but not its human
counterpart, has a differentially methylated CpG island containing
characteristic tandem repeats, which are often found near imprinted
genes.![]() It is thus possible that this island played a role in the evolution of imprinting via enhancement of the expression of Impact.
It is thus possible that this island played a role in the evolution of imprinting via enhancement of the expression of Impact.
Acknowledgements: We thank Genetic Stock Research Center, National Institute of Genetics for JF mice and 14 anonymous individuals for their generous donation of peripheral blood. We are grateful to Tomoyo Shirakawa (University of Tokyo) for helpful discussions. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and from the Japan Society for the Promotion of Science.
 |
 |
 |
 |
 |
 |
 |